“All diseases begin in the gut.”
– Hippocrates (460–370 BC)
In recent years many researchers have described the relationship between the gut microbiota and many diseases, including heart disease, hypertension, diabetes and obesity.1,2 Diet is one of the major factors that influence the pattern of the gut microbiota.3 This article discusses how the gut microbiota affects heart failure.
What is the Human Gut Microbiome?
The collection of micro-organisms that co-exists within or on the host body has been referred to as the microbiota.1 There are more than 2,000 species of commensal organisms (mostly bacteria) that co-exist with the human body, the vast majority in the gut. A healthy human adult has approximately 100 trillion bacteria in the gut, mostly in the colon.1,4
The gut microbiome is acquired from the environment, it is not genetically acquired, and the gut is usually sterile in the womb. For example, the fetus acquires different microbiota during caesarean section and during vaginal delivery.5 Subsequently, the fetus acquires different types of microbiome depending on diet and the environment to which it is exposed.6,7
The human gut microbiome is dominated by five phyla: Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria and Cerrucomicrobia.1,8 Usually the gut microbiota is stable within the individual and family. In the healthy gut, the anaerobic groups Bacteroidetes and Firmicutes contribute to more than 90% of the total bacterial species.8
What Decides the Pattern of an Individual’s Gut Microbiome?
The specific patterns of gut microbiota are called enterotypes.9 An unwelcome change in the gut microbiome is called dysbiosis.10 One of the most important factors that influences the enterotype is the individual’s long‐term diet. For example, diets high in animal protein and fat will show high levels of Bacteroides and low levels of Prevotella (also part of the Bacteroidetes genus).11 On the contrary, diets high in carbohydrates and low in animal protein and fat will have low levels of Bacteroides and high levels of Prevotella. Another example of the diet–gut microbial interaction is found in Japanese people. Their guts contain Bacteroides plebeius, which produces an enzyme that aids in seaweed digestion.12
Other factors that influence the gut microbial pattern other than the diet are environmental changes, hygiene, antibiotic use and disease states.1,6
How the Gut Microbiota Affects the Host
The gut microbiome has many functions.13 One of its functions is a protective function via pathogen displacement, nutrient and receptor competition and production of antimicrobial factors.1 The gut microbiota also secretes some vitamins.
One of the most important functions of the gut microbiome is metabolic, as it aids in the digestion of food components. For example, gut bacteria are involved in the breakdown of sugars (e.g. glycans, which are complex sugars that cannot be cleaved by any human enzymes) by glycoside hydrolase. Gut microbiota participates in the human digestive process through two main catabolic pathways – saccharolytic or proteolytic.14 Both pathways lead to the production of short-chain fatty acids (SCFAs). The second catabolic pathway also produces toxic molecules such as ammonia, various amines, thiols, phenols and indoles, which are cleared by the kidneys but will accumulate if there is renal dysfunction.1,14,15
It is reasonable to view the microbiome as an ‘organ’ that weighs approximately 1–2 kg, although it is without a distinct structure. The microbiome constantly makes compounds, some of which are absorbed and are biologically active. Thus, it can be considered as an endocrine organ producing biologically active entities that diffuse into the bloodstream and act at distant sites.1
The gut microbiota are separated from the lamina propria by a single layer of intestinal epithelium. The intestinal epithelium deploys a variety of mechanisms to restrict commensal bacteria to the intestinal lumen and to prevent egression of these microbiota to the underlying tissue.16 The gut microbiota in turn have evolved to evade the host’s immune system and circumvent the antimicrobial host response.16
The intestinal barrier mechanism has a dual role to play – it protects against the invasion of microorganisms and absorption of bacterial toxins, but also enables the absorption of essential products, electrolytes and nutrients.17
The gut microbiota produces many substances that are able to enter the bloodstream and subsequently influence pathobiological processes. The permeability of these substances is dependent on the functional and structural integrity of the mucosal barrier. Potential barrier disruptors include hypoperfusion of the gut, infections, toxins, drugs and other lifestyle factors.17 Sometimes it may be a structural component of the microbiota itself, such as lipopolysaccharides (LPS) or peptidoglycans, that interact with host mucosal surface cells through pattern recognition receptors.1,18
In addition, molecules produced by microbial organisms can also gain entry to cause various effects. Some identified pathways include the trimethylamine N-oxide (TMAO) pathway, the SCFA pathway and the bile acid pathway.1 The precursor of TMAO is l-carnitine or choline, which is present in food substances such as red meat. If a person has a high intake of red meat, TMAO production is increased, which is implicated in the pathogenesis of heart disease.2
How Do We Study the Gut Microbiome?
It is not easy to study the gut microbiome because it contains millions of bacteria and thousands of species. There are also fungi and viruses present, which can pose difficulties because their genetic material interferes with the identification of the bacterial genome in question. A further issue with studying the gut microbial genome is that the microbial community is distinct in different regions of the intestine, and also because the genome changes frequently due to horizontal gene transfer.19
The traditional method is culture, but it is tedious and time consuming. Bacterial genomic sequencing is the next most suitable method. One popular method is 16S ribosomal RNA (rRNA) gene amplicon analysis. Metagenomic sequencing, another method that is gaining popularity, is usually more expensive but offers increased resolution, enabling a more specific taxonomic and functional classification.20 Wang et al. explained this as: “16S rDNA sequence attempts to reveal ’who’s there?’ in a given microbial community, while shotgun metagenomic sequencing can be used to answer the complementary question of ’what can they do?’.”21
Association of the Gut Microbiota with Heart Disease
There are many recent publications on the association between the gut microbiota and heart disease, especially heart failure.22–26 Changes in the gut microbiota can lead to the development of risk factors for atherosclerotic vascular disease and directly influence pathogenetic disease processes such as acute coronary syndromes and heart failure.27
Obesity is one example. Its pathology is associated with changes in the relative abundance of two dominant bacterial divisions, Bacteroidetes and Firmicutes.28 Obese patients have been shown to display high Firmicutes counts. It has also been found that the obese microbiome has an increased capacity to harvest energy from the diet, and that the obese “trait” is transmissible: colonisation of germ-free mice with an obese microbiota results in a significantly greater increase in total body fat than colonisation with a lean microbiota, with the same diet.29
In addition, hypertension and diabetes have also been found to have associations with specific gut microbial patterns, and researchers have discovered certain links in the pathogenesis of these diseases and bacterial interactions.22,30,31
In a study comparing patients who had coronary heart disease (CHD) with those who did not, it was found that in patients who had CHD, the proportion of the phylum Bacteroidetes was lower, with a higher proportion of Firmicutes.32 Increased TMAO levels were found to be associated with an increased risk of incident major adverse cardiovascular events (MACEs) in a cohort of 4,007 patients who underwent coronary angiography followed up for 3 years.33 In another study, a Cleveland clinic cohort of 530 patients presenting to the emergency department with chest pain showed elevated plasma TMAO levels at presentation that were independently associated with risk of MACEs.34 The Bacteroidetes:Firmicutes ratio is known to be altered in all chronic diseases and therefore may not be a reliable identifier of a particular disease.
Raised TMAO levels are implicated in endothelial and smooth muscle cell activation, foam cell formation, and myocardial and renal fibrosis.2 In a recent systematic review and meta-analysis (16 publications, 19,256 patients), elevated concentrations of TMAO and its precursors were associated with increased risks of MACEs and all-cause mortality, independent of traditional risk factors.35 Another meta-analysis and systematic review of 26,167 patients also showed a positive dose-dependent association between TMAO plasma levels and increased cardiovascular risk and mortality.36
Association of the Microbiota with Heart Failure
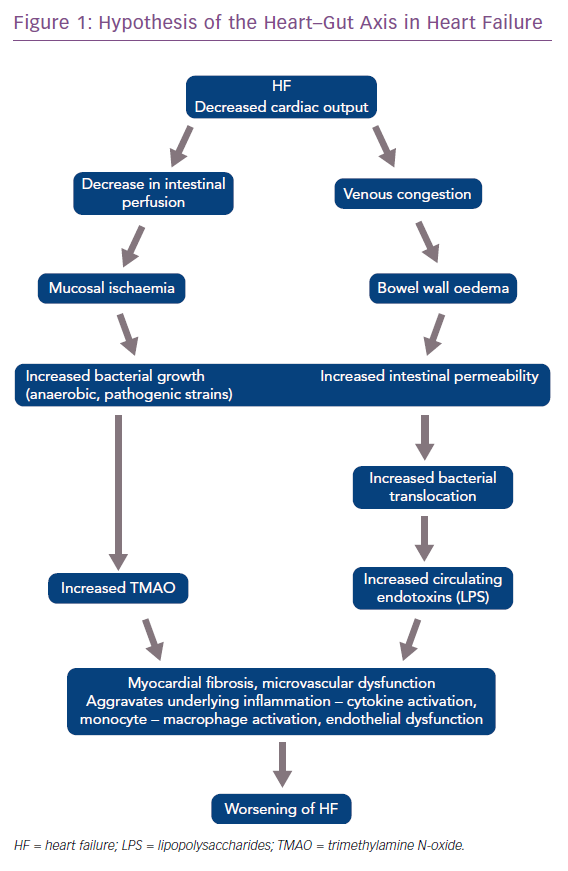
The gut microbiota is also implicated in the pathogenesis of heart failure (HF). In HF, due to reduced ejection fraction, there is a reduction in intestinal blood flow and low oxygen delivery. This predisposes the gut to the growth of pathogenic types of anaerobic bacteria.37 Patients with chronic HF also develop bowel wall oedema due to venous congestion that impedes the absorptive function of the gut and permits bacterial overgrowth in the mucus layer adjacent to the apical surface of the colonic mucosa.36 Increased intestinal permeability, assessed by the sugar cellobiose test, has also been reported in patients with HF, and this increased permeability correlates with right atrial pressure and C-reactive protein levels.38,39
These bacteria produce many harmful substances including TMAO and endotoxin (LPS), which predisposes or leads to worsening of HF. These discoveries have led to the hypothesis of the heart–gut axis of HF (Figure 1).40,41 Higher LPS concentrations have been found in patients with decompensated HF, which correlates with the increased level of bowel wall oedema, as discussed earlier. LPS decreases after ‘re-compensation’. According to Sandek et al., this suggests a cause and effect relationship between the oedematous gut wall, epithelial dysfunction and translocating LPS.42
High TMAO levels are found in patients with HF, which predict higher long-term mortality, even after adjusting for traditional risk factors and cardiorenal indexes.41 TMAO has been found to be a prognostic factor in HF patients, and higher levels predict a poor prognosis at 1-year follow-up. A combination of TMAO and the traditional marker N-terminal pro-brain natriuretic peptide are able to provide additional prognostic information.43
Why do TMAO levels increase to such an extent in HF? The changes in bacterial composition, as discussed earlier, appear to be the primary driver of TMAO levels.25 Renal impairment and changing dietary patterns may also contribute.25 How TMAO affects the pathobiology of HF is not clear. Proposed theories include stimulation of cytokines such as tumour necrosis factor-alpha, which can aggravate myocardial fibrosis, microvascular dysfunction in the heart independent of its proatherosclerotic effects, neurohormonal derangements, and so on, but we do not yet have a clear answer.25
Can We Manipulate the Gut Microbiome to Treat Disease?
There are some studies on manipulation of the gut microbiome that give us hope in treating related diseases. Manipulation can be achieved in many ways. We can alter the diet to change the type of microbiota, we can target the chemicals produced by the gut microbiota, or we can directly alter the microbial flora by the addition of probiotics.
If we reduce red meat in the diet, we reduce the intake of choline and lecithin, and thereby reduce TMAO, which has a positive impact on the risk of heart disease. For example, changing to a Mediterranean diet has been shown to reduce markers of HF. Another method is to administer nonabsorbable antibiotics that kill specific microbiota and thus alter the overall microbial pattern.
Probiotics is another method that can alter the gut’s microbial pattern. Probiotics are live beneficial bacteria (Bifidobacteria, Lactobacilli, Streptococci and non-pathogenic strains of Escherichia coli) that can be ingested to create an appropriate intestinal microbial balance. There are studies using Saccharomyces boulardii in HF that have shown benefit. However, the positive effects of probiotics only apply to a restricted group of microbial species and potential hazards exist, including the possibility of turning these microbiota into opportunistic pathogens in immunocompromised individuals.44
The ongoing Gut-Heart trial has randomised 150 patients with stable HF and a left ventricular ejection fraction <40% to receive the antibiotic rifaximin, the probiotic yeast S boulardii (ATCC 74012) or no treatment in a 1:1:1 fashion.45 The primary endpoint is ejection fraction at 3 months. The outcome of the trial will shed some light into the possible therapeutic avenues in the future targeting gut microbiome.
The last – and very interesting – method that is gaining popularity in the treatment of many gastrointestinal diseases is faecal transplantation. Faecal transplantation from lean volunteers was found to show a benefit in weight reduction as well as a reduction in risk factor levels for HF.46
We are not yet sure of the best method to alter the gut microbiota; however, the most safe and promising option may be to rely on alteration of the diet.
Conclusion
Millions of years of co-evolution have created diverse ecosystems of gut microbiota that contribute to the maintenance of human metabolic homeostasis. We are slowly discovering the various ways that these co-habitants work in health and disease. We are therefore not alone – we are linked with our gut microbiota, which controls our systems remotely. Understanding and manipulating the microbiota may hold future answers for health and disease.